Seventh competition following Youhan Lee's curriculum. Object segmentation(Deep learning) competition.
2018 Data Science Bowl
Find the nuclei in divergent images to advance medical discovery
www.kaggle.com
First Kernel: Teaching notebook for total imaging newbies
- "This kernel will implement classical image techniques and will hopefully serve as a useful primer to people who have never worked with image data before"
Insight / Summary:
1. Removing background
- Basic Concept of Background Removal
- The image is divided into two classes:
- Objects of interest
- Background
- This approach assumes that image pixel intensities follow a bimodal distribution
- A bimodal distribution means a distribution with two peaks
- The image is divided into two classes:
- Threshold Setting Methods:
- Basic Statistical Methods:
- Using mean: Uses the average of all pixel values as threshold
- Using median: Uses the middle value of all pixel values as threshold
- Disadvantage: May not effectively reflect actual image characteristics
- Otsu Method:
- Working Principle:
- Calculates variance between two classes (object and background) for all possible thresholds
- Selects threshold that maximizes between-class variance
- Simultaneously minimizes within-class variance
- Advantages:
- Automatically finds optimal threshold
- Particularly effective for bimodal distributions
- Computationally efficient
- Characteristics:
- Histogram-based method
- Global thresholding method (applies same threshold across entire image)
- This method is particularly effective for processing medical images and microscope images where there is a clear brightness difference between the background and objects.
- Working Principle:
- Basic Statistical Methods:
from skimage.filters import threshold_otsu
thresh_val = threshold_otsu(im_gray)
mask = np.where(im_gray > thresh_val, 1, 0)
# Make sure the larger portion of the mask is considered background
if np.sum(mask==0) < np.sum(mask==1):
mask = np.where(mask, 0, 1)
2. Deriving individual masks for each object
- For this contest, we need to get a separate mask for each nucleus.
- One way we can do this is by looking for all objects in the mask that are connected, and assign each of them a number using ndimage.label.
- Then, we can loop through each label_id and add it to an iterable, such as a list.
from scipy import ndimage
labels, nlabels = ndimage.label(mask)
label_arrays = []
for label_num in range(1, nlabels+1):
label_mask = np.where(labels == label_num, 1, 0)
label_arrays.append(label_mask)
print('There are {} separate components / objects detected.'.format(nlabels))
# Result: There are 76 separate components / objects detected.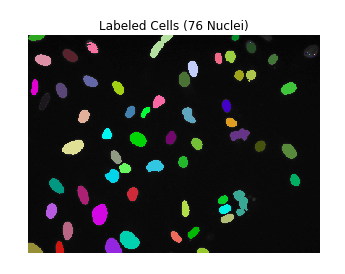
Detailed Explanation:
This code shows the process of separating individual objects (in this case, nuclei) from an image and creating masks for each.
1. `ndimage.label()` function explanation:
labels, nlabels = ndimage.label(mask)- Groups connected pixels in a binary mask
- Returns:
- `labels`: Array where each pixel is assigned a label number
- `nlabels`: Total number of objects found
- Example:
Input mask: Label result:
1 1 0 0 1 1 1 1 0 0 2 2
1 1 0 0 1 1 → 1 1 0 0 2 2
0 0 1 1 0 0 0 0 3 3 0 0
2. Creating individual object masks:
label_arrays = []
for label_num in range(1, nlabels+1):
label_mask = np.where(labels == label_num, 1, 0)
label_arrays.append(label_mask)- For each label number (1 to nlabels):
- Uses `np.where(condition, value_if_true, value_if_false)`
- Sets pixels equal to current label number to 1
- Sets all other pixels to 0
- Results in separate binary masks for each object
3. Checking results:
print('There are {} separate components / objects detected.'.format(nlabels))- Prints total number of objects found
Real example:
# Original binary mask
mask = np.array([
[1, 1, 0, 0, 1, 1],
[1, 1, 0, 0, 1, 1],
[0, 0, 1, 1, 0, 0]
])
# After applying ndimage.label()
labels = [
[1, 1, 0, 0, 2, 2],
[1, 1, 0, 0, 2, 2],
[0, 0, 3, 3, 0, 0]
]
# Mask for first object
label_mask_1 = [
[1, 1, 0, 0, 0, 0],
[1, 1, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0]
]
- This process is particularly important in cell image analysis.
- It's necessary when analyzing each nucleus individually or measuring characteristics like size and shape.
3. Problems
- A quick glance reveals two problems (in this very simple image):
- There are a few individual pixels that stand alone (e.g. top-right)
- Some cells are combined into a single mask (e.g., top-middle)
- Using ndimage.find_objects, we can iterate through our masks, zooming in on the individual nuclei found to apply additional processing steps.
- find_objects returns a list of the coordinate range for each labeled object in your image.
4. Removing too small labels:
for label_ind, label_coords in enumerate(ndimage.find_objects(labels)):
cell = im_gray[label_coords]
# Check if the label size is too small
if np.product(cell.shape) < 10:
print('Label {} is too small! Setting to 0.'.format(label_ind))
mask = np.where(labels==label_ind+1, 0, mask)
# Regenerate the labels
labels, nlabels = ndimage.label(mask)
print('There are now {} separate components / objects detected.'.format(nlabels))
Result:
Label 4 is too small! Setting to 0.
Label 5 is too small! Setting to 0.
Label 7 is too small! Setting to 0.
Label 8 is too small! Setting to 0.
Label 9 is too small! Setting to 0.
Label 10 is too small! Setting to 0.
Label 14 is too small! Setting to 0.
Label 15 is too small! Setting to 0.
Label 16 is too small! Setting to 0.
Label 19 is too small! Setting to 0.
Label 21 is too small! Setting to 0.
Label 22 is too small! Setting to 0.
Label 23 is too small! Setting to 0.
Label 60 is too small! Setting to 0.
Label 61 is too small! Setting to 0.
Label 72 is too small! Setting to 0.
There are now 60 separate components / objects detected.Detailed Explanation:
if np.product(cell.shape) < 10:
print('Label {} is too small! Setting to 0.'.format(label_ind))
mask = np.where(labels==label_ind+1, 0, mask)
- np.product(cell.shape): Calculates number of pixels in object
- If less than 10 pixels:
- Prints the label
- Removes the object from mask (sets to 0)
Example:
# Original labels
[
[1, 1, 0, 2], # Label 2 is a single pixel
[1, 1, 0, 0],
[0, 0, 3, 0]
]
# After size check (Label 2 removed)
[
[1, 1, 0, 0], # Small label 2 is removed
[1, 1, 0, 0],
[0, 0, 3, 0]
]This process helps to keep only meaningful nuclei by removing very small objects that might be due to noise or artifacts.
5. Adjacent cell problem
fig, axes = plt.subplots(1,6, figsize=(10,6))
for ii, obj_indices in enumerate(ndimage.find_objects(labels)[0:6]):
cell = im_gray[obj_indices]
axes[ii].imshow(cell, cmap='gray')
axes[ii].axis('off')
axes[ii].set_title('Label #{}\nSize: {}'.format(ii+1, cell.shape))
plt.tight_layout()
plt.show()
- Label #2 has the "adjacent cell" problem: the two cells are being considered part of the same object.
- One thing we can do here is to see whether we can shrink the mask to "open up" the differences between the cells.
- This is called mask erosion.
- We can then re-dilate it to recover the original proportions.
# Get the object indices, and perform a binary opening procedure
two_cell_indices = ndimage.find_objects(labels)[1]
cell_mask = mask[two_cell_indices]
cell_mask_opened = ndimage.binary_opening(cell_mask, iterations=8)Details:
This code performs morphological operations to separate attached cells:
- ndimage.find_objects(labels)[1]:
- [1] selects the second label (Label #2)
- This retrieves coordinates of an area containing two attached cells
- mask[two_cell_indices]:
- Extracts only the mask of the selected area
- This area contains the two attached cells
- ndimage.binary_opening():
- Performs binary opening operation
- Opening consists of two steps:
- Erosion: Erodes object boundaries inward
- Dilation: Restores eroded objects back to original size
- iterations=8:
- Repeats erosion and dilation 8 times
- More iterations result in stronger separation effect
Original mask: After erosion: After dilation:
1 1 1 1 0 0 0 0 1 0 0 1
1 1 1 1 → 0 1 1 0 → 1 1 1 1
1 1 1 1 0 0 0 0 1 0 0 1Effects of this process:
- Erosion breaks thin connections between cells
- Dilation restores the main parts of cells to original size
- Results in separation of previously attached cells
This is an important preprocessing step in cell image analysis. It's necessary to analyze individually cells that were initially attached to each other.
Result:
fig, axes = plt.subplots(1,4, figsize=(12,4))
axes[0].imshow(im_gray[two_cell_indices], cmap='gray')
axes[0].set_title('Original object')
axes[1].imshow(mask[two_cell_indices], cmap='gray')
axes[1].set_title('Original mask')
axes[2].imshow(cell_mask_opened, cmap='gray')
axes[2].set_title('Opened mask')
axes[3].imshow(im_gray[two_cell_indices]*cell_mask_opened, cmap='gray')
axes[3].set_title('Opened object')
for ax in axes:
ax.axis('off')
plt.tight_layout()
plt.show()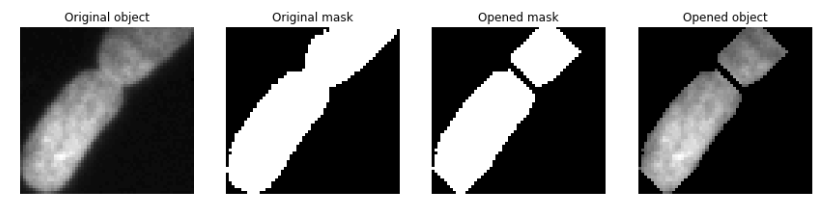
6. Convert each labeled object to Run Line Encoding
- Finally, we need to encode each label_mask into a "run line encoded" string.
- Basically, we walk through the array, and when we find a pixel that is part of the mask, we index it and count how many subsequent pixels are also part of the mask.
- We repeat this each time we see new pixel start point.
def rle_encoding(x):
'''
x: numpy array of shape (height, width), 1 - mask, 0 - background
Returns run length as list
'''
dots = np.where(x.T.flatten()==1)[0] # .T sets Fortran order down-then-right
run_lengths = []
prev = -2
for b in dots:
if (b>prev+1): run_lengths.extend((b+1, 0))
run_lengths[-1] += 1
prev = b
return " ".join([str(i) for i in run_lengths])
print('RLE Encoding for the current mask is: {}'.format(rle_encoding(label_mask)))RLE Encoding for the current mask is: 210075 6 210593 8 211112 9 211631 10 212150 11 212669 12 213189 12 213709 12 214228 13 214748 13 215268 13 215788 13 216308 13 216828 13 217348 13 217869 12 218389 12 218909 12 219430 11 219950 11 220471 10 220991 10 221512 9 222033 8 222554 7 223075 6 223596 5 224117 4 224639 2Details:
Advantages of this encoding:
- Data compression: Represents consecutive same values only with starting point and length
- Efficient storage: Especially saves storage space in large images
- Fast transmission: Can transmit data in compressed form
The output result "210075 6 210593 8 ..." means:
- 6 pixels starting from position 210075
- 8 pixels starting from position 210593 and so on.
Example:
1) Original image (2x8 size):
0 1 1 0 0 1 1 1
0 0 1 1 0 0 1 02) After transpose:
0 0
1 0
1 1
0 1
0 0
1 0
1 1
1 0- Rows and columns are swapped
- Now read from top to bottom, then right
3) After flattening:
[0 0 1 0 1 1 0 0 1 0 1 1 1 1 1 0]
- Transposed array arranged in a line
- Index: 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
4) RLE encoding "3 1 5 2 9 1 11 5":
0 0 [1 0] 1 1 0 0 1 0 1 1 1 1 1 0
^
1 from position 30 0 1 0 [1 1] 0 0 1 0 1 1 1 1 1 0
^
2 from position 50 0 1 0 1 1 0 0 [1] 0 1 1 1 1 1 0
^
1 from position 90 0 1 0 1 1 0 0 1 0 [1 1 1 1 1] 0
^
5 from position 11
7. Combine it into a single function
- Now that we've seen the basic steps to processing an image in a "dumb" way, we can combine it all into a single function.
- This function will take an image path, perform the processes outlined above, and spit out a dataframe with the RLE strings for each mask found.
- We also create a wrapper function that will spit out a single DataFrame for all images in the dataset.
import pandas as pd
def analyze_image(im_path):
'''
Take an image_path (pathlib.Path object), preprocess and label it, extract the RLE strings
and dump it into a Pandas DataFrame.
'''
# Read in data and convert to grayscale
im_id = im_path.parts[-3]
im = imageio.imread(str(im_path))
im_gray = rgb2gray(im)
# Mask out background and extract connected objects
thresh_val = threshold_otsu(im_gray)
mask = np.where(im_gray > thresh_val, 1, 0)
if np.sum(mask==0) < np.sum(mask==1):
mask = np.where(mask, 0, 1)
labels, nlabels = ndimage.label(mask)
labels, nlabels = ndimage.label(mask)
# Loop through labels and add each to a DataFrame
im_df = pd.DataFrame()
for label_num in range(1, nlabels+1):
label_mask = np.where(labels == label_num, 1, 0)
if label_mask.flatten().sum() > 10:
rle = rle_encoding(label_mask)
s = pd.Series({'ImageId': im_id, 'EncodedPixels': rle})
im_df = im_df.append(s, ignore_index=True)
return im_df
def analyze_list_of_images(im_path_list):
'''
Takes a list of image paths (pathlib.Path objects), analyzes each,
and returns a submission-ready DataFrame.'''
all_df = pd.DataFrame()
for im_path in im_path_list:
im_df = analyze_image(im_path)
all_df = all_df.append(im_df, ignore_index=True)
return all_dftesting = pathlib.Path('../input/stage1_test/').glob('*/images/*.png')
df = analyze_list_of_images(list(testing))
df.to_csv('submission.csv', index=None)Second Kernel: Keras U-Net starter - LB 0.277
- Shows how to get started on segmenting nuclei using a neural network in Keras
Insight / Summary:
1. U-Net
- The architecture used is the so-called U-Net, which is very common for image segmentation problems such as this.
- I believe they also have a tendency to work quite well even on small datasets.
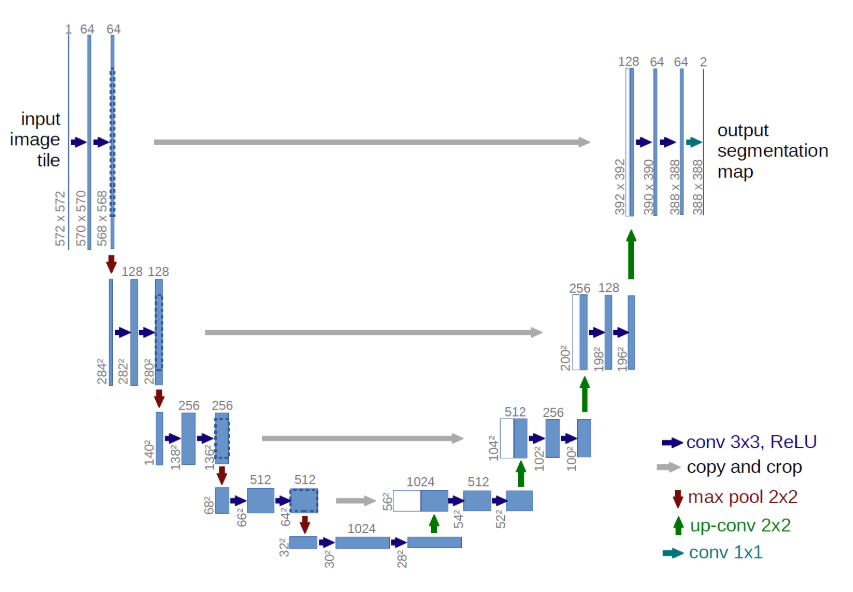
2.
- We fit the model on the training data, using a validation split of 0.1.
- We use a small batch size because we have so little data.
- I recommend using checkpointing and early stopping when training your model.
Third Kernel:Nuclei Overview to Submission
- the preprocessing steps to load the data
- a quick visualization of the color-space
- training a simple CNN
- applying the model to the test data
- creating the RLE test data
Success is not about being better than others, but about being better than you were yesterday.
- Max Holloway -
'캐글' 카테고리의 다른 글
| [Kaggle Study] #10 Zillow Prize: Zillow’s Home Value Prediction (Zestimate) (0) | 2024.11.29 |
|---|---|
| [Kaggle Study] #9 New York City Taxi Trip Duration (0) | 2024.11.29 |
| [Kaggle Study] #6 Costa Rican Household Poverty Level Prediction (1) | 2024.11.28 |
| [Kaggle Study] #7 TensorFlow Speech Recognition Challenge (0) | 2024.11.27 |
| [Kaggle Study] #4 More about Home Credit Default Risk Competition (0) | 2024.11.26 |